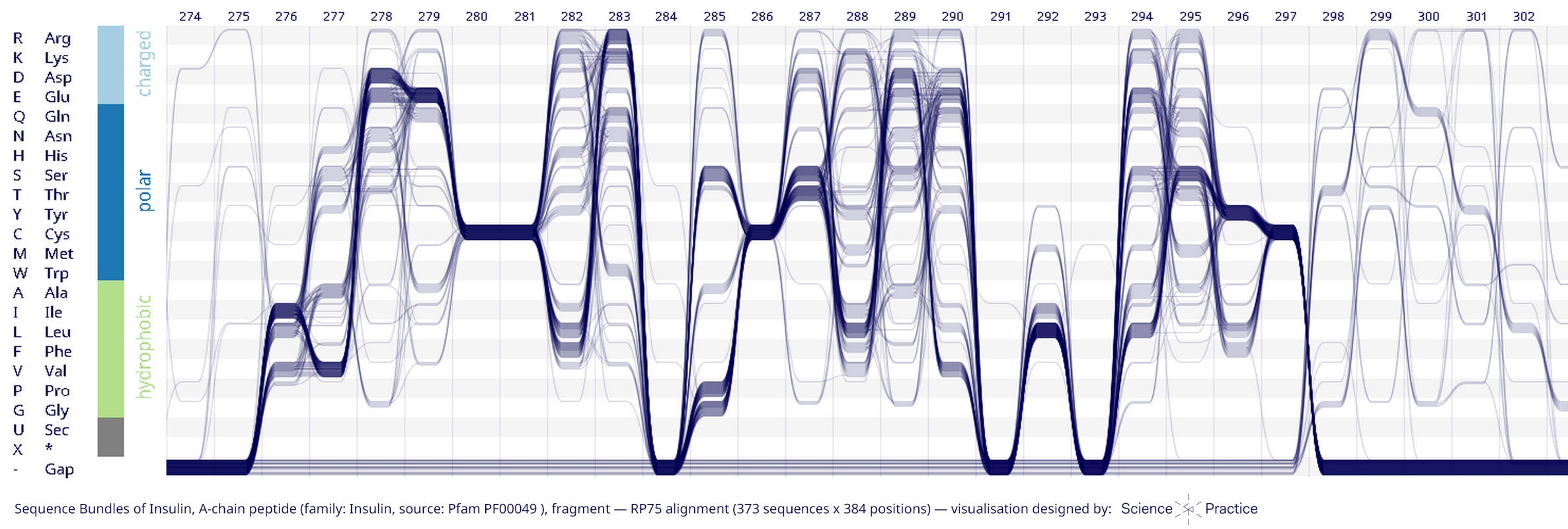
INSULIN
Insulin is a protein hormone involved in the management and use of glucose in the organism. Key parts of insulin are universally conserved across all organisms — this includes six Cysteines in positions 90, 102, 280, 281, 286, 297, responsible for forming structurally important sulphur bridges.
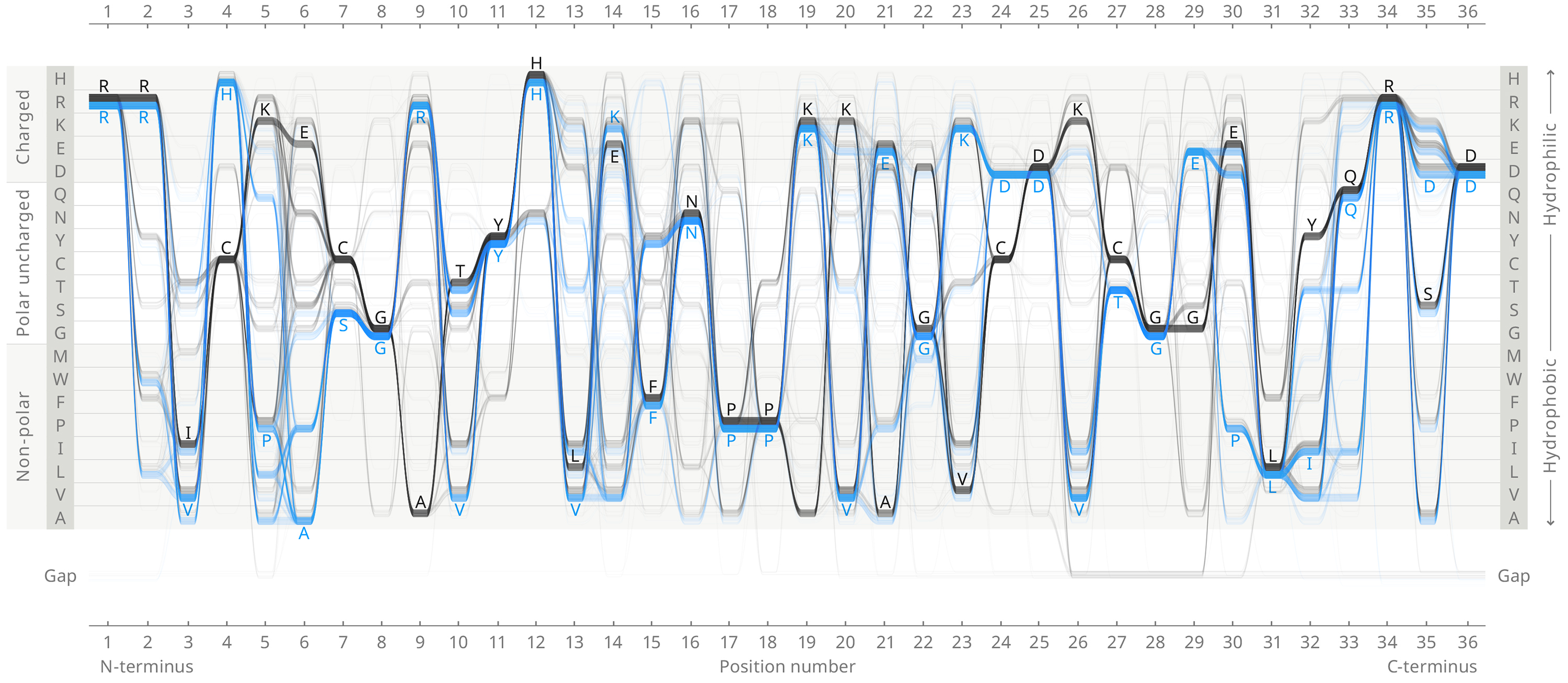
ADK_LID
The ADK_LID domain structure is universally conserved, but is stabilized in the Gram-negatives (blue sequences) by a hydrogen bonding network between residues 4, 7, 9, 24, 27, and 29 (and several other residues in some organisms), while the Gram-positives (black sequences) are stabilized by a bound metal ion, tetrahedrally coordinated by the Cysteines at 4, 7, 24 and 27.
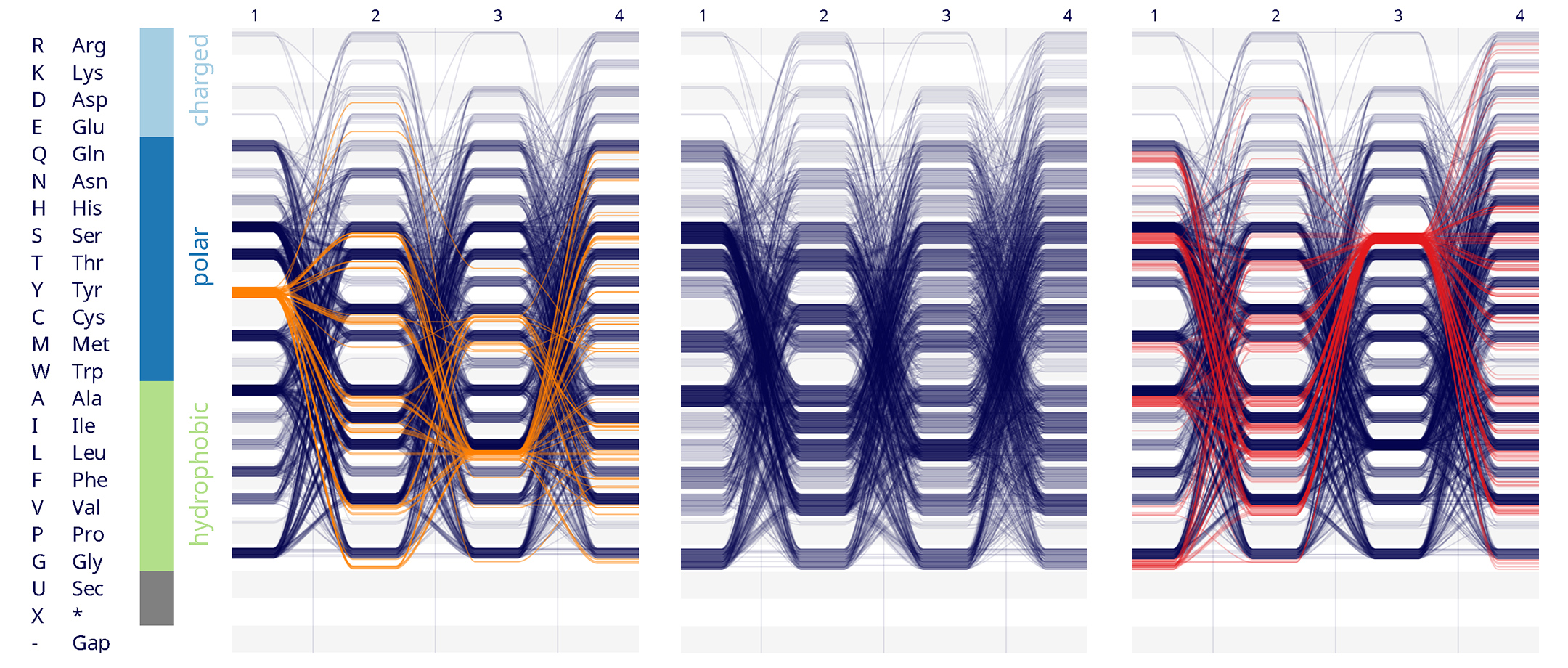
HK:RR class of signalling protein
Histidine Kinase:Response Regulator (HK:RR) is a class of simple signalling proteins that have been well characterised and can be re-engineered for different applications e.g. to create biosensor bacteria. Here, four important positions on the HK (which determine binding specifically to its partner, the RR) were combinatorially
mutated to 160,000 unique 4-amino-acid-long sequences. Out of these 160,000 only 1,659 have been found to be functional. We visualised this list of 1,659 sequences using Sequence Bundles to provide a salient representation of the pattern responsible for allowing the HK:RR protein interaction to work.
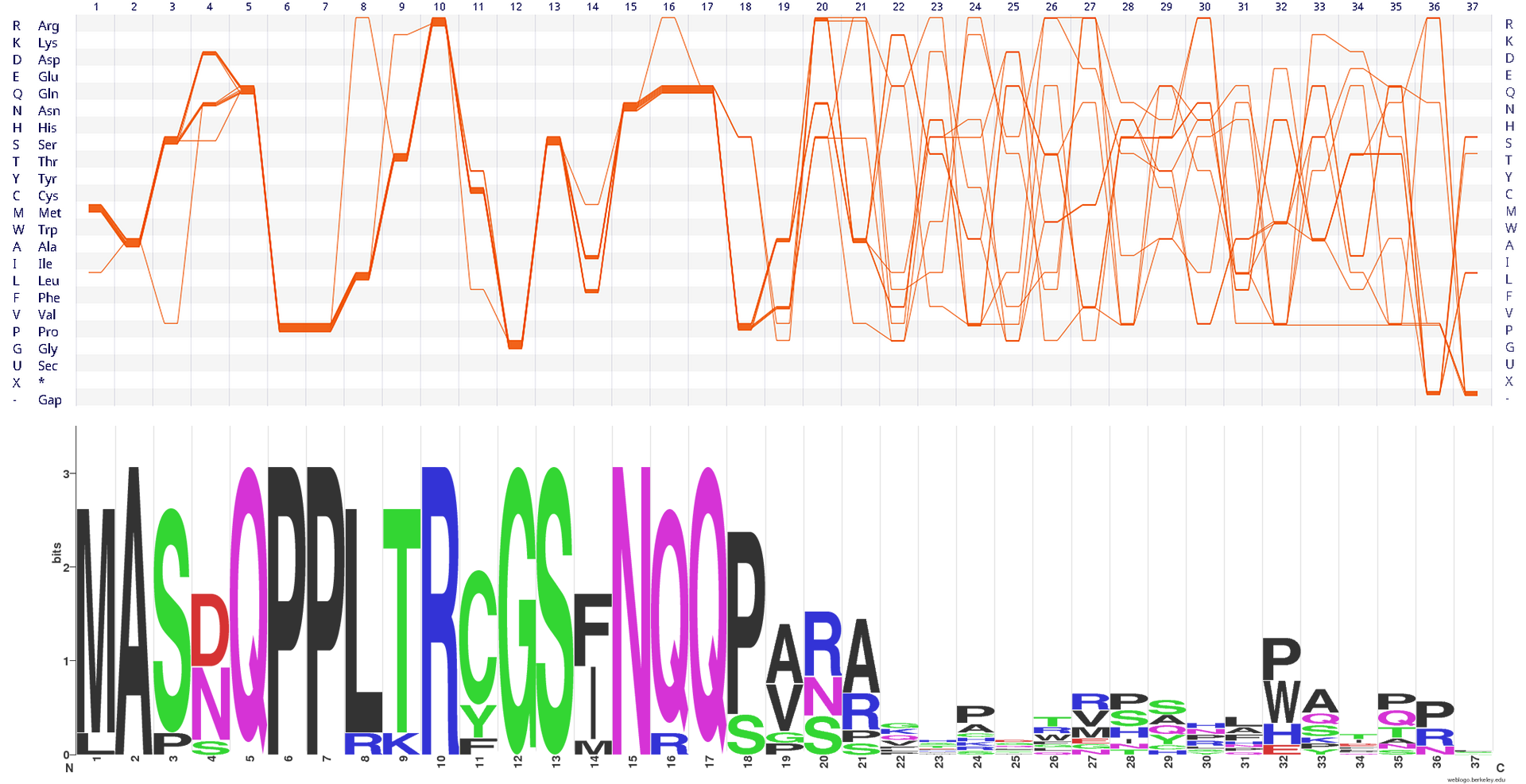
QPP motif in Rhodopirellula baltica
The QPP motif is a very small alignment containing 11 sequences, each
37 positions long. We used the full Pfam PF07640 alignment. The QPP proteins are found in Rhodopirullela baltica, and they share a strongly conserved fragment centred around the invariant QPP motif (positions 5–7) located at their N termini (left part of the visualisation).
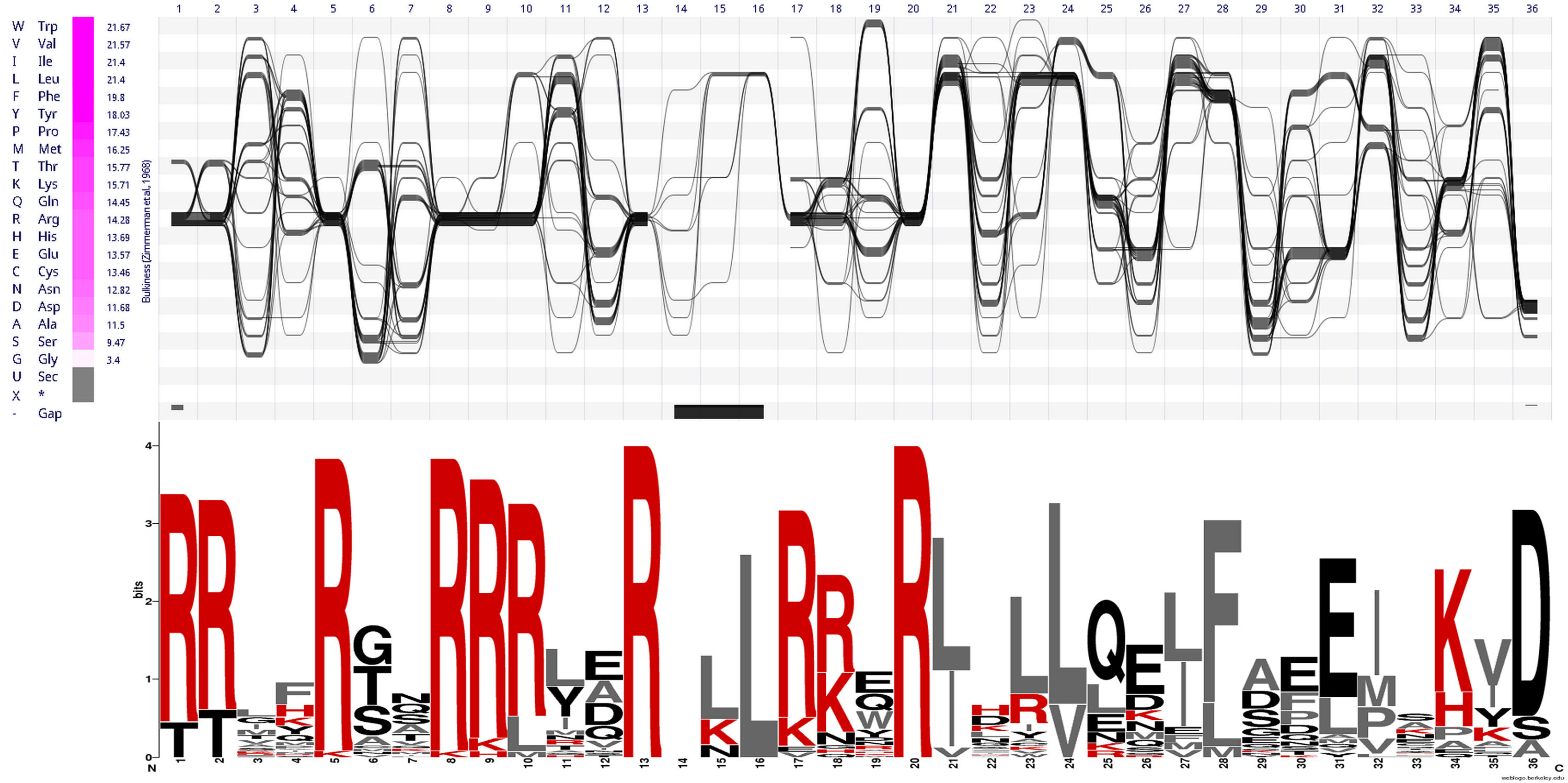
CRISPR Cas9 bridge helix
CRISPR Cas9 enabled a revolutionary gene editing technology that allows
very precise changes to be made in the DNA code of living cells. CRISPR Cas9 brings new capability to biosciences, but also novel risks, which are only now being addressed by the community. The CRISPR Cas9 gene editing protein comprises three key elements: two lobes (NUC lobe and REC lobe) and a bridge helix which connects both lobes. The bridge helix is rich in highly conserved arginines (R) and facilitates both Cas9 lobes to precisely target those DNA sections that will be edited.